The Future of CRISPR Therapeutics for Human Hematopoietic Stem Cell Derived Diseases
by Courtney Gegg, MS | July 12, 2018
CRISPR gene editing enables curative autologous treatments for hematopoietic stem cell-derived diseases
Using CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) to genetically engineer hematopoietic stem cells (HSCs) for therapeutic applications is an emerging market that holds promise for the treatment of HSC-derived diseases. HSCs reside in peripheral blood and bone marrow, divide indefinitely to provide a limitless HSC pool, and differentiate into every type of blood cell [1]. In practice, an ex vivo CRISPR-directed HSC therapy involves the following steps:
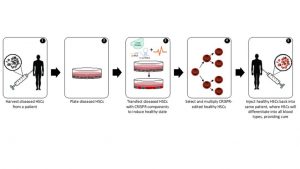
Figure 1. Ex vivo CRISPR editing process.
The CRISPR-mediated genome engineering process modifies genetic code by introducing breaks at targeted locations within a genome. These breaks are then fixed through the cell’s endogenous repair processes, which can insert or delete one or more nucleotides to inactivate or replace a gene.
The CRISPR HSC gene editing market for monogenic HSC diseases is nascent, but companies are beginning to enter to compete for the ~$1B annual market
Current state-of-the-art genome editing technologies include zinc finger nucleases and transcription activator-like effector nucleases. These technologies are time-consuming, expensive, difficult to multiplex to target multiple genomic loci, and require technical expertise to engineer [2]. In contrast, CRISPR technology is fast to engineer, affordable, easily multiplexed, and user-friendly [2]. Due to these advantages, CRISPR has rapidly gained the largest segment of the genome editing market [3], despite being the youngest technology.
HSC CRISPR therapies are most easily suited to treat diseases caused by a mutation in a single gene, also known as monogenic disorders. There are 10,000 monogenic diseases [4], affecting about 6% of people [5]. Initial CRISPR HSC therapeutics are targeting monogenic blood-related diseases that can be treated through simple gene deletion or inactivation achieved through nonhomologous end-joining (NHEJ) (Figure 2) [6]. Diseases that require gene correction are more difficult to treat as they rely on homology directed recombination (HDR) (Figure 2). HDR is about one-third as efficient as NHEJ in proliferating human cells, which means that harnessing NHEJ is a better strategy for human therapeutics [7].
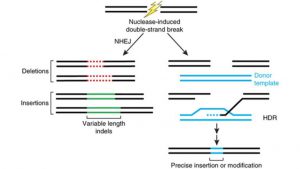
Figure 2. Endogenous repair mechanisms employed by cells once a double-stranded break is initiated: Non-homologous end joining which results in gene deletion or inactivation (left) and homology directed recombination which results in gene replacement (right). Image from [8].
Monogenic blood-related diseases treated through simple gene deletion or inactivation include thalassemia, sickle cell anemia, severe combined immunodeficiency, and hemophilia [4]. There are 1000, 1550, 70, and 1000 Americans born annually with these diseases, respectively [9-12]. The only curative treatment for these diseases is an allogenic HSC transplant, in which cells in a diseased patient’s bloodstream are eradicated and repopulated with healthy HSCs from a donor. These procedures cost around $275,000 [13] and often require lifelong immunosuppressants, which cost around $12,000 per year per patient [14, 15]. Therefore, the annual market for these diseases is currently around $1B. Autologous CRISPR-edited HSC transplants, which involve harvesting cells from a patient, editing those cells, and injecting the healthy cells back into the same patient are more attractive than allogenic transplants, as they cost about half as much and do not require immunosuppressants [16].
There are currently three main companies in the CRISPR autologous HSC therapeutic space: Intellia Therapeutics, CRISPR Therapeutics, and Editas Medicine. These companies are beginning with ex vivo CRISPR approaches targeting beta-thalassemia and sickle cell disease, but in vivo CRISPR applications are in their pipelines [17-19].
Academic research is beginning to surmount technical limitations of CRISPR
CRISPR technologies are currently limited by their low efficiency and specificity. Efficiency is a measurement of how often the target sequence is edited and specificity is a measurement of how often off-target bases are changed. Current low efficiency of on-target mutations (typically <10%) must be overcome before widespread use [20]. High off-target activity results in unwanted modifications and is the biggest threat to the CRISPR-Cas9 technology. Off-target activity ranges from ~2-35% and has been measured in some instances to be greater than the on-target rate [20-22].
Strategies to increase efficiency increase HDR over NHEJ, as NHEJ outcompetes HDR in mammalian cells, limiting the therapeutic potential of CRISPR to treat diseases that require gene correction
By synchronizing the expression of Cas9 with phases of the cell cycle in which HDR is most active, the rate of HDR can be increased up to ~90% [23, 24]. Additionally, HDR rates are improved though the use of single-stranded DNA donors that asymmetrically align with the double-stranded break created by Cas9 [25]. It is also possible to repress key NHEJ molecules through shRNAs or small molecules such that HDR is dominant; this leads to an 8-fold increase of HDR [26]. Lastly, homology-independent targeted integration [27] uses NHEJ to allow gene knock-ins, resulting in gene integration at target sites roughly 10-fold higher than traditional HDR [28].
Truncated guide RNAs, paired nicking strategy, and fusion of Cas9 with FokI reduce off-target effects by orders of magnitude compared to wild-type Cas9
Guide RNAs with shorter regions of complementarity to the target loci can decrease off-target mutations by up to 5000-fold [29]. Pairing a mutant nickase version of Cas9 that makes single-strand breaks in DNA with a pair of offset sgRNAs can reduce off-target activity by 50- to 1,500 fold [30]. Fusion of FokI nuclease to a variant of Cas9 that requires association of two fCas9 monomers to initiate cleavage modifies DNA target sites with 140-fold higher specificity [31, 32].
Recent academic breakthroughs that improve base pair editing specificity increase utility of CRISPR technology
Base editing is the modification of single ribonucleic acid bases and leads to increased efficiency and specificity above the current system. Recently, a research group demonstrated the ability to convert A-T base pairs to G-C base pairs with 50% efficiency and low rates of indels (<0.1%). Though this technology can only edit A-T to G-C bases, more than half of point mutations, which account for >60% of all genetic mutations that lead to human disease, are caused by a change from G-C to A-T [33]. Another group later achieved precise A-to-I RNA edits. RNA editing is transient, rendering it less dangerous than DNA editing and useful for temporary disease states such as inflammation [34]. Base editing technologies will be used to complement traditional CRISPR technology, rather than replace.
New immunity knowledge brings attention to development of additional Cas9 variants
In a 2018 study pre-print, researchers claim that >65% of healthy patients have antibodies against the two most commonly used bacterial Cas9 proteins. The significance of this is unknown, though it may suggest that humans may be able to inactivate the CRISPR/Cas9 system [35]. However, engineering Cas9 to make it less detectable by the immune system or using Cas9 proteins from organisms not known to infect humans could easily address this problem.
While technical limitations are being addressed, other barriers still exist that deter entry into the CRISPR market
Intellectual Property Landscape: Since 2016, two founding CRISPR factions have been engaged in a patent dispute. This uncertainty deters many new potential entrants from developing CRISPR-based products as it is unclear who can to properly license this technology[36].
Regulatory Considerations: As CRISPR gene editing is in its infancy, initial technology platform discussions with the FDA could help determine the possibility of approving CRISPR for certain indications as well as the regulatory pathway CRISPR companies will need to follow for approval.
Uncertain Reimbursement Structure: Under the 2010 Affordable Care Act, charging more or refusing to cover individuals with “pre-existing conditions” (which include HSC-related congenital genomic diseases) was prohibited [37]. However, whether this patient protection will remain in current proposed legislation is uncertain [38]. Furthermore, because CRISPR-mediated gene therapies are a one-time, curative treatment, the traditional insurance model of multiple small payments over a set time period is ill-fit [39].
Ethical Considerations: Even if CRISPR editing for human therapeutics is approved by the FDA, it may be rejected in the marketplace due to negative consumer perception. In 2014, half of Americans said they would reject gene editing on themselves or their children, even for reducing risk of developing serious diseases [40].
CRISPR technology is a promising gene editing technology with the potential to revolutionize the gene editing market once technical limitations are addressed
CRISPR technology is user-friendly, fast to engineer, affordable, and can be multiplexed, rendering it advantageous over preceding gene editing techniques. Nevertheless, the barriers to entry into CRISPR HSC therapies are high due to the intellectual property landscape, regulatory aspects, reimbursement structure, ethical considerations, and most essentially, technological limitations. However, innovation is beginning to successfully surmount the technological limitations, leaving the CRISPR HSC therapeutic space primed for a breakthrough.
References
[1] M.N.I.o.H. NIH Stem Cell Information Home Page. In Stem Cell Information [World Wide Web site]. Bethesda, U.S. Department of Health and Human Services, 2016 [cited February 8, 2018] Available at < //stemcells.nih.gov/info/Regenerative_Medicine/2006Chapter4.htm>.
[2] T. Gaj, C.A. Gersbach, C.F. Barbas, ZFN, TALEN and CRISPR/Cas-based methods for genome engineering, Trends in biotechnology 31(7) (2013) 397-405.
[3] Kalorama Information, Gene Editing Markets, 2016.
[4] World Health Organization, Genes and human disease: Monogenic diseases. <http://www.who.int/genomics/public/geneticdiseases/en/index2.html>, 2018).
[5] A.S. Rodwell C., eds., “2014 Report on the State of the Art of Rare Disease Activities in Europe”, July 2014.
[6] W.-J. Dai, L.-Y. Zhu, Z.-Y. Yan, Y. Xu, Q.-L. Wang, X.-J. Lu, CRISPR-Cas9 for in vivo Gene Therapy: Promise and Hurdles, Molecular Therapy – Nucleic Acids 5.
[7] Z. Mao, M. Bozzella, A. Seluanov, V. Gorbunova, Comparison of nonhomologous end joining and homologous recombination in human cells, DNA Repair 7(10) (2008) 1765-1771.
[8] J.D. Sander, J.K. Joung, CRISPR-Cas systems for editing, regulating and targeting genomes, Nature biotechnology 32(4) (2014) 347-55.
[9] Bluebirdbio, Patients & Families: B-thalassemia. <https://www.bluebirdbio.com/patients-families/beta-thalassemia/>).
[10] CDC, Sickle Cell Disease (SCD): Data and Statistics. <https://www.cdc.gov/ncbddd/sicklecell/data.html>).
[11] CDC, Addition of Severe Combined Immunodeficiency as a Contraindication for Administration of Rotavirus Vaccine, 2010.
[12] National Hemophilia Foundation, Fast Facts: About Bleeding Disorders. <https://www.hemophilia.org/About-Us/Fast-Facts>, 2018).
[13] M.S. Broder, T.P. Quock, E. Chang, S.R. Reddy, R. Agarwal-Hashmi, S. Arai, K.F. Villa, The Cost of Hematopoietic Stem-Cell Transplantation in the United States, American Health & Drug Benefits 10(7) (2017) 366-374.
[14] T.F. Page, R.S. Woodward, Cost of Lifetime Immunosuppression Coverage for Kidney Transplant Recipients, Health Care Financing Review 30(2) (2008) 95-104.
[15] B.L. Kasiske, D. Cohen, M.R. Lucey, J.F. Neylan, Payment for immunosuppression after organ transplantation. American Society of Transplantation, Jama 283(18) (2000) 2445-50.
[16] N.S. Majhail, L.-W. Mau, E.M. Denzen, T.J. Arneson, Costs of Autologous and Allogeneic Hematopoietic Cell Transplantation in the United States: A Study Using a Large National Private Claims Database, Bone marrow transplantation 48(2) (2013) 294-300.
[17] C.C. Analytics, Intellia Therapeutics Inc – Cortellis Company Detailed Pipeline Report, 2018.
[18] Cortellis Clarivate Analysis, CRISPR Therapeutics AG – Cortellis Company Detailed Pipeline Report Philidelphia, PA, 2018.
[19] Cortellis Clarivate Analytics, Editas Medicine Inc – Cortellis Company Detailed Pipeline Report, Cortellis Clarivate Analytics, 2018.
[20] L. Cong, F.A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P.D. Hsu, X. Wu, W. Jiang, L.A. Marraffini, F. Zhang, Multiplex Genome Engineering Using CRISPR/Cas Systems, Science (New York, N.Y.) 339(6121) (2013) 819-823.
[21] Y. Fu, J.A. Foden, C. Khayter, M.L. Maeder, D. Reyon, J.K. Joung, J.D. Sander, High frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells, Nature biotechnology 31(9) (2013) 822-826.
[22] P.D. Hsu, D.A. Scott, J.A. Weinstein, F.A. Ran, S. Konermann, V. Agarwala, Y. Li, E.J. Fine, X. Wu, O. Shalem, T.J. Cradick, L.A. Marraffini, G. Bao, F. Zhang, DNA targeting specificity of RNA-guided Cas9 nucleases, Nature biotechnology 31(9) (2013) 827-32.
[23] S. Lin, B.T. Staahl, R.K. Alla, J.A. Doudna, Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery, eLife 3 (2014) e04766.
[24] T. Gutschner, M. Haemmerle, G. Genovese, G.F. Draetta, L. Chin, Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair, Cell reports 14(6) (2016) 1555-1566.
[25] C.D. Richardson, G.J. Ray, M.A. DeWitt, G.L. Curie, J.E. Corn, Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA, Nature biotechnology 34(3) (2016) 339-44.
[26] V.T. Chu, T. Weber, B. Wefers, W. Wurst, S. Sander, K. Rajewsky, R. Kuhn, Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells, Nature biotechnology 33(5) (2015) 543-8.
[27] S.S. De Ravin, L. Li, X. Wu, U. Choi, C. Allen, S. Koontz, J. Lee, N. Theobald-Whiting, J. Chu, M. Garofalo, C. Sweeney, L. Kardava, S. Moir, A. Viley, P. Natarajan, L. Su, D. Kuhns, K.A. Zarember, M.V. Peshwa, H.L. Malech, CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease, Science translational medicine 9(372) (2017).
[28] K. Suzuki, Y. Tsunekawa, R. Hernandez-Benitez, J. Wu, J. Zhu, E.J. Kim, F. Hatanaka, M. Yamamoto, T. Araoka, Z. Li, M. Kurita, T. Hishida, M. Li, E. Aizawa, S. Guo, S. Chen, A. Goebl, R.D. Soligalla, J. Qu, T. Jiang, X. Fu, M. Jafari, C.R. Esteban, W.T. Berggren, J. Lajara, E. Nunez-Delicado, P. Guillen, J.M. Campistol, F. Matsuzaki, G.H. Liu, P. Magistretti, K. Zhang, E.M. Callaway, K. Zhang, J.C. Belmonte, In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration, Nature 540(7631) (2016) 144-149.
[29] Y. Fu, J.D. Sander, D. Reyon, V.M. Cascio, J.K. Joung, Improving CRISPR-Cas nuclease specificity using truncated guide RNAs, Nature biotechnology 32(3) (2014) 279-284.
[30] F.A. Ran, Patrick D. Hsu, C.-Y. Lin, Jonathan S. Gootenberg, S. Konermann, A.E. Trevino, David A. Scott, A. Inoue, S. Matoba, Y. Zhang, F. Zhang, Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity, Cell 154(6) 1380-1389.
[31] J.P. Guilinger, D.B. Thompson, D.R. Liu, Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification, Nature biotechnology 32(6) (2014) 577-582.
[32] S.Q. Tsai, N. Wyvekens, C. Khayter, J.A. Foden, V. Thapar, D. Reyon, M.J. Goodwin, M.J. Aryee, J.K. Joung, Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing, Nature biotechnology 32(6) (2014) 569-76.
[33] N.M. Gaudelli, A.C. Komor, H.A. Rees, M.S. Packer, A.H. Badran, D.I. Bryson, D.R. Liu, Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage, Nature 551(7681) (2017) 464-471.
[34] D.B.T. Cox, J.S. Gootenberg, O.O. Abudayyeh, B. Franklin, M.J. Kellner, J. Joung, F. Zhang, RNA editing with CRISPR-Cas13, Science (New York, N.Y.) 358(6366) (2017) 1019.
[35] C.T. Charlesworth, P.S. Deshpande, D.P. Dever, B. Dejene, N. Gomez-Ospina, S. Mantri, M. Pavel-Dinu, J. Camarena, K.I. Weinberg, M.H. Porteus, Identification of Pre-Existing Adaptive Immunity to Cas9 Proteins in Humans, bioRxiv (2018).
[36] Piper Jaffray Companies, CRISPR Therapeutics (CRSP) Transformative Technology with Huge Therapeutic Potential; Assume at Overweight, (2017).
[37] Patient Protection and Affordable Care Act, 42 U.S.C. § 18001 et seq. (2010).
[38] J.S. Sherkow, CRISPR, Patents, and the Public Health (December 17, 2017). Yale Journal of Biology and Medicine, vol. 90, pp. 667-672 (2017); NYLS Legal Studies Research Paper No. 3093610. Available at SSRN: https://ssrn.com/abstract=3093610.
[39] Spark Therapeutics, Spark Therapeutics Announces First-of-their-kind Programs to Improve Patient Access to LUXTURNA™ (voretigene neparvovec-rzyl), a One-time Gene Therapy Treatment, 2018.
[40] J. Pew Research Center, 2015, “Americans, Politics and Science Issues”.


