
Trends and Opportunities in Antibiotic Development for Multidrug-Resistant Bacterial Infections
by Maria Filsinger Interrante, BS | July 12, 2018
Antimicrobial resistance is major global health concern and is insufficiently addressed by the current clinical pipeline
The advent of antibiotic therapy was one of the most impactful medical innovations of the 20th century. However, as resistance has waxed and antibiotic development has waned, multidrug-resistant bacterial infections have been increasingly recognized as a global health crisis of staggering proportions, with recent estimates predicting that annual deaths due to drug-resistant bacteria will reach 10 million worldwide by 2050 [1].
Why has antibiotic discovery, development, and approval lagged? A combination of market forces which include high regulatory hurdles, short and curative treatment courses, low prices, the threat of resistance, and appropriately conservative “last-line” use by physicians has made antibiotic development less economically attractive to pharmaceutical companies than pursuing other indications [2]. Furthermore, it is challenging to enroll seriously ill patients in clinical trials when they require urgent treatment and while the infectious agent responsible for their disease may still be unknown. To make matters worse, two Supreme Court cases in 2012 and 2013 have made it harder to patent natural products (the vast majority of currently-approved antibiotics are natural products), potentially prohibiting companies from securing exclusivity on the sale of their products. The impact of these factors is dramatic: the number of large U.S. and European pharmaceutical companies doing antimicrobial research shrank from 36 in 1980 to just 4 in 2016 [3], with the number of new FDA approvals of antibiotics falling precipitously during the same time period.
It comes as no surprise, then, that in late 2017 the WHO deemed the current clinical pipeline “insufficient to mitigate the threat of antimicrobial resistance,” citing an urgent need for new classes of antibiotics with little or no cross-resistance to existing drugs [4]. Innovation, investment, and entrepreneurship in the antibiotic space has never been more critical. This article will provide a high-level summary of current trends in antibiotic development for multidrug-resistant pathogens, identify key areas of opportunity for growth, and provide a framework for evaluating companies and assets in the space.
The clinical antibiotics pipeline is inappropriately skewed towards redevelopment or combination of existing drugs, exploitation of existing mechanisms of action and chemical matter, and neglects to target many of the highest-priority pathogens
In late 2017 the WHO released a comprehensive, 48-page analysis of all antibacterial agents in clinical development [4]. While efforts incentivizing basic research and early trials have swollen the pipeline somewhat, a closer examination of the 51 small molecules and 11 biologicals (representing 42 new therapeutic entities) reveals several critical weaknesses and gaps:

Outside of the limitations noted by the WHO, existing incentive programs channel push funding the earliest stages of pharmaceutical development, leaving small- and medium-sized enterprises without the capital needed to advance through late stage clinical trials and make it to market. In addition, large pharma companies tend to not be incentivized by push funding at all, and the lack of pull incentives make the cost of acquiring an early-stage asset and pushing it through to market a large and unappealing risk.
Surveying the clinical landscape with the above gaps in mind, a handful of companies to watch in the coming years are Medimmune (with two monoclonal antibodies in phase 2 clinical trials, one targeting Staph aureus and one targeting Pseudomonas aeruginosa), Contrafect and Intron (both of which are advancing phage endolysins into phase 1 and 2a trials, respectively), Polyphor (with an novel, membrane targeting IV antibiotic in phase 2 trials against Pseudomonas aeruginosa), and Debiopharm (with a Fabl inhibitor dosed both orally and IV in phase 2 trials against Staph aureus). While targeting new mechanisms of action with novel chemical matter is undoubtedly more risky in the short term than making a small modification to an existing drug, successful drugs will likely evade resistance development for significantly longer, as well as be capable of treating patients for which every other possible pharmaceutical intervention has been exhausted.
While preclinical antibiotic development has been bolstered by recent incentives and features several new and promising therapeutic approaches, a new discovery engine and improved testing methods are required for a robust and sustainable pipeline in the long term
The increasing recognition of multidrug-resistant bacterial infections as a global health crisis and the aforementioned incentives that this realization triggered have undoubtedly moved the needle in preclinical antibiotic development. While most agents already in clinical trials utilize existing chemotypes or target well-exploited mechanisms, some of the more original innovations in antibiotic therapy are still in the preclinical arena [Figure 1, 10].
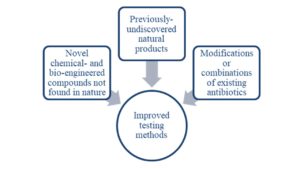
Previously-undiscovered or unused natural products
Figure 1. Sources of new antibiotics likely to be effective against multi-drug resistant pathogens.
One “natural product” (here meant as an unmodified agent derived from nature) that has recently been re-examined as a promising approach to defeating MDR pathogens is phage therapy, in which whole live phages (viruses that usually infect bacteria) are used to kill pathogens and thus cure infection. In 2016, a patient with multidrug-resistant A. baumannii was successfully treated with IV phage therapy at UC San Diego under an emergency investigational new drug application from the FDA [11], further fueling interest in developing phage approaches for the clinic. One company pursuing this approach is AmpliPhi Biosciences, with a 3-phage combination therapy for S. aureus having completed Phase I trials and a second, 4-phage combination therapy for P. aeruginosa available for certain patients via expanded axess. A second company, Adaptive Phage Therapeutics, seeks to combine phage technology with AI and sequencing to determine the best combination of phages for a given patient’s infection, and has also treated several patients through expanded access. That said, phage therapy still faces significant challenges in production, formulation, and FDA approval due to the heterogeneity of the product and the difficulty in standardizing and mass-producing such a therapy.
On the in silico side, advances in computational biology have been leveraged has been to mine genomic data in search of both new bacterial targets and new natural product antibiotics. One standout player in this sphere is Warp Drive Bio, a Cambridge company that secured a $750M R&D collaboration with Sanofi in 2016 and another $387M collaboration with Roche in 2017. With an assembled genomic database of over 135,000 strains, a proprietary “genomic search engine,” and a “genomes to molecules” synthetic biology platform, Warp Drive Bio has built a powerful discovery engine capable of accessing novel antibiotic classes. This approach, which synthesizes several key advances in genomics, computer science, and synthetic biology, is exactly the kind of integrated engine necessary to meet the demand for new antibiotics – entrepreneurs and investors alike should take note.
Another exciting potential discovery engine, this time that leverages hardware instead of software, is the iChip device. Essentially a plastic array of small diffusion chambers, iChip enables the “domestication” of previously bacterial species that could not be cultured, many of which produce antimicrobial metabolites, a handful of which have demonstrated early promise as novel antibiotics [16]. While exciting, this chip-based method and others like it that rely on culturing bacteria and extracting, purifying, and testing natural products face significant challenges in scale-up that genome-based methods do not [17]. The fate of NovoBiotic, a company whose founders include the inventor of iChip and that has using iChip for antibiotic discovery for the past 5 years, will be an excellent indication of the promise of this approach.
Modifications or combinations of existing antibiotics
An approach borrowed from recent developments in the field of oncology involves antibody-drug conjugates to more selectively deliver an antibiotic payload to bacteria while sparing host cells. Recently, Genentech developed an antibody-antibiotic conjugate against S. aureus that indeed improved the pharmacokinetics of the antibacterial and was able to reduce the bacterial load in a mouse model of systemic S. aureus infection after a single dose [13]. Antibody-antibiotic conjugate development is significantly de-risked by the existence and approval of antibody-drug conjugates for other indications, though the relatively low potency of antibiotics compared to chemotherapeutics and the variable expression levels of the bacterial antigens will be unique challenges to overcome. Also of note are siderophore-drug conjugates and engineered bacteriophage endolyins, both of which have already advanced to clinical trials.
Chemical- or bio-engineered compounds not found in nature
Recently, in silico tools have begun to show promise for generating novel antibiotics through increasingly sophisticated modeling and docking software. In 2015, Iscla et. al. built a spatial map of the exposed oxygen atoms of amino acids lining the gate of their target, the MscL channel, then used iterative docking models to screen and optimize ligands. One compound they pulled out, dubbed Ramizol, was capable of curing MRSA infections in S. aureus and belongs to a new class of antimicrobials [14]. The compound was further tested in rodent infection models and was found to have low systemic absorption, a favorable pharmacokinetic profile, and showed promising survival data in a C. difficile hamster model [15]. The success of this approach highlights the potential for in silico design tools to be applied to antibiotic development, especially in the service of accessing fundamentally new chemotypes.
Current methods utilized to determine antibiotic susceptibility must be critically examined and in many cases, re-engineered to address biofilm infections
While the importance of discovery engines for novel antibiotics cannot be overstated, an essential piece of the translation from a newly-discovered molecule to a lead compound advancing through clinical trials is the methodology used to determine antimicrobial activity. Current methods for determining minimum inhibitory concentration (MIC) for an antimicrobial compound involve susceptibility testing in which homogenous, planktonic organisms float freely in suspension to which an antibiotic is added. However, this model has recently been called into question as the most appropriate method for assessing susceptibility in the face of growing awareness of biofilms and the role that they play in infection [18]. Indeed, 80% of microbial infections in humans are biofilm-mediated, and organisms growing in biofilms as opposed to in suspension have required up to a 1000-fold increase in antibiotic concentration to achieve the same effects [19]. In fact, the relative disassociation between planktonic susceptibility and therapeutic success may be due in large part to misdirected screening that ignores the role of biofilms in human colonization.
While certainly more complex than planktonic susceptibility testing, biofilm susceptibility screens can and should become the standard for assessing antimicrobial activity in light of the critical role that biofilms play in most multidrug-resistant infections which these agents are intended to clinically address. In addition to the importance of new agents being tested in these models, there is reason to believe that there are compounds in pharmaceutical libraries that were passed-over in planktonic screens but may be effective in biofilms and indeed be promising clinical candidates. If we are seeking compounds capable of killing heterogeneous, matrix-embedded, adhered organisms in residing in multi-species communities, then this is what should be used in our screening approaches going forward.
Governments, NGOs, and public-private collaborations have developed programs to bolster antibiotic discovery in academia and industry
Recognizing the need for acceleration in antibiotic development, governments, NGOs, and public-private partnerships have begun rolling out grants and initiatives. For example, in 2016 WHO and the Drugs for Neglected Diseases initiative launched the Global Antibiotic Research and Development Partnership (GARDP) to prioritize pathogens, fund research programs, and expand access, and in 2016 the US government partnered with the Welcome Trust to create CARB-X, a non-profit that selects and funds pre-clinical research [5, 6]. In addition, governments have stepped up to incentivize innovation, with a pertinent example in the United States being the passage of the FDA GAIN (Generating Antibiotic Incentives Now) Act in 2012, which enables sponsors to request a “Qualified Infectious Disease Product” (QIDP) designation for drugs addressing antibiotic-resistant infections, granting an additional 5 years of patent exclusivity in addition to eligibility for fast track designation and priority review. These efforts and others provide strong push incentives for basic research and early clinical trials, helping to make antibiotic development an attractive business model once again [7]. Potential investors should also note that drugs with indications in infectious disease were doubly as likely to reach approval from phase I (19.1% compared to 9.6% and 25.2% compared to 13.8%, by two recent estimates) as the overall, indicating that investment in antibiotic lead candidates is actually less risky from a likelihood of approval standpoint than many other pharmaceutical areas [8,9].
Given both the critical unmet medical need and the developing infrastructure to incentivize and support antibiotic development, now is a time of significant opportunity for both entrepreneurs and investors in this space (Table 2).

References
[1] – Antimicrobial Resistance: Global Report on Surveillance, WHO, 2014
http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf
[2] – C. Lee Ventola, The Antibiotic Resistance Crisis Pt 1: Causes and Threats https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4378521/
[3] – Andrew Chung, Yasmeen Abutaleb, Deborah J. Nelson, As ‘superbugs’ strengthen, an alarming lack of new weapons to fight them, 2016
https://www.reuters.com/investigates/special-report/usa-uncounted-drugs/,
[4] – Antibacterial Agents in Clinical Development, WHO, 2017 http://apps.who.int/iris/bitstream/10665/258965/1/WHO-EMP-IAU-2017.11-eng.pdf?ua=1
[5] – Global Antibiotic Research and Development Project, 2016
https://www.gardp.org/
[6] – CARB-X
http://www.carb-x.org/
[7] – V L Simpkin, M J Renwick, R Kelly, E Mossialos, Incentivizing innovation in antibiotic drug discovery and development: progress, challenges and next steps, The Journal of Antibiotics volume 70, pages 1087–1096 (2017)
https://www.nature.com/articles/ja2017124
[8] BIO, Biomedtracker, Amplion, Clinical Development Success Rates, 2006-2015 (June 2016)
https://www.bio.org/sites/default/files/Clinical%20Development%20Success%20Rates%202006-2015%20-%20BIO,%20Biomedtracker,%20Amplion%202016.pdf
[9] Chi Heem Wong, Kien Wei Siah, Andrew W Lo; Estimation of clinical trial success rates and related parameters, Biostatistics, kxx069 (2018)
https://doi.org/10.1093/biostatistics/kxx069
[10] – A Mullard, Preclinical antibiotic pipeline gets a pick-me-up, Nature Reviews Drug Discovery 16, 741–742 (2017)
https://www.nature.com/articles/nrd.2017.213
[11] – Bacteriophage therapy treats patient near death with MDR Acinetobacter baumannii, Outbreak News Today, April 2017 http://outbreaknewstoday.com/bacteriophage-therapy-treats-patient-near-death-mdr-acinetobacter-baumannii-45488/
[12] – S Mariathasan, MW Tan, Antibody-Antibiotic Conjugates: A Novel Therapeutic Platform against Bacterial Infections, Trends Mol Med. 2017 Feb;23(2):135-149
http://www.cell.com/trends/molecular-medicine/pdf/S1471-4914(16)30193-9.pdf
[13] – C Zhou, S Lehar, J Gutierrez, CM Rosenberger, N Ljumanovic, J Dinoso, N Koppada, K Hong, A Baruch, M Carrasco-Triguero, O Saad, S Mariathasan & AV Kamath (2016) Pharmacokinetics and pharmacodynamics of DSTA4637A: A novel THIOMAB™ antibody antibiotic conjugate against Staphylococcus aureus in mice, mAbs, 8:8, 1612-1619
http://www.tandfonline.com/doi/full/10.1080/19420862.2016.1229722
[14] – I Iscla, R Wray, P Blount, J Larkins-Ford, AL Conery, FM Ausubel, S Ramu, A Kavanagh, JX Huang, MA Blaskovich, MA Cooper, A Obregon-Henao, I Orme, ES Tjandra, UH Stroeher, MH Brown, C Macardle, NV Holst, CL Tong, AD Slattery, CT Gibson, CL Raston and RA Boulos A new antibiotic with potent activity targets MscL, The Journal of Antibiotics (2015) 68, 453–462
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4430313/
[15] – S Rao, CA Prestidge, L Miesel, D Sweeney, DL Shinabarger, R Boulos, Preclinical development of Ramizol, an antibiotic belonging to a new class, for the treatment of Clostridium difficile colitis, The Journal of Antibiotics volume 69, pages 879–884 (2016)
https://www.ncbi.nlm.nih.gov/pubmed/27189122
[16] – E Gavrish, CS Sit, S Cao, O Kandror, A Spoering, A Peoples, L Ling, A Fetterman, D Hughes, A Bissell, H Torrey, T Akopian, A Mueller, S Epstein, A Goldberg, J Clardy, K Lewis. Lassomycin, a Ribosomally Synthesized Cyclic Peptide, Kills Mycobacterium tuberculosis by Targeting the ATP-Dependent Protease ClpC1P1P2, Chemistry & Biology, Volume 21, Issue 4, 24 April 2014, Pages 509-518
https://www.sciencedirect.com/science/article/pii/S1074552114000763
[17] – A Katsnelson, New Twist on Antibiotic Hunt Hits Pay Dirt, Nature Reviews Drug Discovery 14, 153–154 (2015)
https://www.nature.com/articles/nrd4556
[18] – JN Pendleton; SP Gorman; BF Gilmore, Clinical Relevance of the ESKAPE Pathogens, Expert Rev Anti Infect Ther. 2013;11(3):297-308.
https://www.medscape.com/viewarticle/780768_11
[19] – Howard Ceri, Merle E Olson & Raymond J Turner (2010) Needed, new paradigms in antibiotic development, Expert Opinion on Pharmacotherapy, 11:8, 1233-1237
http://www.tandfonline.com/doi/full/10.1517/14656561003724747


